二十种特征变换方法及Spark MLlib调用实例(Scala/Java/python)(二)
2016-11-30 17:24
866 查看
VectorIndexer
算法介绍:
VectorIndexer解决数据集中的类别特征Vector。它可以自动识别哪些特征是类别型的,并且将原始值转换为类别指标。它的处理流程如下:
1.获得一个向量类型的输入以及maxCategories参数。
2.基于原始数值识别哪些特征需要被类别化,其中最多maxCategories需要被类别化。
3.对于每一个类别特征计算0-based类别指标。
4.对类别特征进行索引然后将原始值转换为指标。
索引后的类别特征可以帮助决策树等算法处理类别型特征,并得到较好结果。
在下面的例子中,我们读入一个数据集,然后使用VectorIndexer来决定哪些特征需要被作为非数值类型处理,将非数值型特征转换为他们的索引。
调用示例:
Scala:
Java:
Python:
Normalizer(正则化)
算法介绍:
Normalizer是一个转换器,它可以将多行向量输入转化为统一的形式。参数为p(默认值:2)来指定正则化中使用的p-norm。正则化操作可以使输入数据标准化并提高后期学习算法的效果。
下面的例子展示如何读入一个libsvm格式的数据,然后将每一行转换为
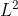
以及
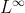
形式。
调用示例:
Scala:
Java:
Python:
StandardScaler
算法介绍:
StandardScaler处理Vector数据,标准化每个特征使得其有统一的标准差以及(或者)均值为零。它需要如下参数:
1. withStd:默认值为真,使用统一标准差方式。
2. withMean:默认为假。此种方法将产出一个稠密输出,所以不适用于稀疏输入。
StandardScaler是一个Estimator,它可以fit数据集产生一个StandardScalerModel,用来计算汇总统计。然后产生的模可以用来转换向量至统一的标准差以及(或者)零均值特征。注意如果特征的标准差为零,则该特征在向量中返回的默认值为0.0。
下面的示例展示如果读入一个libsvm形式的数据以及返回有统一标准差的标准化特征。
调用示例:
Scala:
Java:
Python:
MinMaxScaler
算法介绍:
MinMaxScaler通过重新调节大小将Vector形式的列转换到指定的范围内,通常为[0,1],它的参数有:
1. min:默认为0.0,为转换后所有特征的下边界。
2. max:默认为1.0,为转换后所有特征的下边界。
MinMaxScaler计算数据集的汇总统计量,并产生一个MinMaxScalerModel。该模型可以将独立的特征的值转换到指定的范围内。
对于特征E来说,调整后的特征值如下:
%20=%20%5Cfrac%7B%7B%7Be_i%7D%20-%20%7BE_%7B%5Cmin%20%7D%7D%7D%7D%7B%7B%7BE_%7B%5Cmax%20%7D%7D%20-%20%7BE_%7B%5Cmin%20%7D%7D%7D%7D*(%5Cmax%20%20-%20%5Cmin%20)%20+%20%5Cmin)
如果

,则
)
。
注意因为零值转换后可能变为非零值,所以即便为稀疏输入,输出也可能为稠密向量。
下面的示例展示如果读入一个libsvm形式的数据以及调整其特征值到[0,1]之间。
调用示例:
Scala:
Java:
Python:
MaxAbsScaler
算法介绍:
MaxAbsScaler使用每个特征的最大值的绝对值将输入向量的特征值转换到[-1,1]之间。因为它不会转移/集中数据,所以不会破坏数据的稀疏性。
下面的示例展示如果读入一个libsvm形式的数据以及调整其特征值到[-1,1]之间。
调用示例:
Scala:
Java:
Python:
Bucketizer
算法介绍:
Bucketizer将一列连续的特征转换为特征区间,区间由用户指定。参数如下:
1. splits:分裂数为n+1时,将产生n个区间。除了最后一个区间外,每个区间范围[x,y]由分裂的x,y决定。分裂必须是严格递增的。在分裂指定外的值将被归为错误。两个分裂的例子为Array(Double.NegativeInfinity,0.0,
1.0, Double.PositiveInfinity)以及Array(0.0, 1.0, 2.0)。
注意,当不确定分裂的上下边界时,应当添加Double.NegativeInfinity和Double.PositiveInfinity以免越界。
下面将展示Bucketizer的使用方法。
调用示例:
Scala:
Java:
Python:
ElementwiseProduct
算法介绍:
ElementwiseProduct按提供的“weight”向量,返回与输入向量元素级别的乘积。即是说,按提供的权重分别对输入数据进行缩放,得到输入向量v以及权重向量w的Hadamard积。
*%5Cleft(%20%5Cbegin%7Barray%7D%7Bl%7D%7Bw_1%7D%5C%5C...%5C%5C%7Bw_n%7D%5Cend%7Barray%7D%20%5Cright)%20=%20%5Cleft(%20%5Cbegin%7Barray%7D%7Bl%7D%7Bv_1%7D%7Bw_1%7D%5C%5C...%5C%5C%7Bv_n%7D%7Bw_n%7D%5Cend%7Barray%7D%20%5Cright))
下面例子展示如何通过转换向量的值来调整向量。
调用示例:
Scala:
Java:
Python:
SQLTransformer
算法介绍:
SQLTransformer工具用来转换由SQL定义的陈述。目前仅支持SQL语法如"SELECT
...FROM __THIS__ ...",其中"__THIS__"代表输入数据的基础表。选择语句指定输出中展示的字段、元素和表达式,支持Spark SQL中的所有选择语句。用户可以基于选择结果使用Spark
SQL建立方程或者用户自定义函数。SQLTransformer支持语法示例如下:
1. SELECTa, a + b AS a_b FROM __THIS__
2. SELECTa, SQRT(b) AS b_sqrt FROM __THIS__ where a > 5
3. SELECTa, b, SUM(c) AS c_sum FROM __THIS__ GROUP BY a, b
示例:
假设我们有如下DataFrame包含id,v1,v2列:
id | v1 | v2
----|-----|-----
0 | 1.0 | 3.0
2 | 2.0 | 5.0
使用SQLTransformer语句"SELECT *,(v1 + v2) AS v3, (v1 * v2) AS v4 FROM __THIS__"转换后得到输出如下:
id | v1 | v2 | v3 | v4
----|-----|-----|-----|-----
0 | 1.0| 3.0 | 4.0 | 3.0
2 | 2.0| 5.0 | 7.0 |10.0
调用示例:
Scala:
Java:
Python:
VectorAssembler
算法介绍:
VectorAssembler是一个转换器,它将给定的若干列合并为一列向量。它可以将原始特征和一系列通过其他转换器得到的特征合并为单一的特征向量,来训练如逻辑回归和决策树等机器学习算法。VectorAssembler可接受的输入列类型:数值型、布尔型、向量型。输入列的值将按指定顺序依次添加到一个新向量中。
示例:
假设我们有如下DataFrame包含id,hour,mobile,
userFeatures以及clicked列:
id | hour | mobile| userFeatures | clicked
----|------|--------|------------------|---------
0 |18 | 1.0 | [0.0, 10.0, 0.5] | 1.0
userFeatures列中含有3个用户特征。我们想将hour,mobile以及userFeatures合并为一个新列。将VectorAssembler的输入指定为hour,mobile以及userFeatures,输出指定为features,通过转换我们将得到以下结果:
id | hour | mobile| userFeatures | clicked | features
----|------|--------|------------------|---------|-----------------------------
0 |18 | 1.0 | [0.0, 10.0, 0.5] | 1.0 | [18.0, 1.0, 0.0, 10.0, 0.5]
调用示例:
Scala:
Java:
Python:
QuantileDiscretizer
算法介绍:
QuantileDiscretizer讲连续型特征转换为分级类别特征。分级的数量由numBuckets参数决定。分级的范围有渐进算法决定。渐进的精度由relativeError参数决定。当relativeError设置为0时,将会计算精确的分位点(计算代价较高)。分级的上下边界为负无穷到正无穷,覆盖所有的实数值。
示例:
假设我们有如下DataFrame包含id,hour:
id | hour
----|------
0 |18.0
----|------
1 |19.0
----|------
2 | 8.0
----|------
3 | 5.0
----|------
4 | 2.2
hour是一个Double类型的连续特征,将参数numBuckets设置为3,我们可以将hour转换为如下分级特征。
id | hour | result
----|------|------
0 |18.0 | 2.0
----|------|------
1 |19.0 | 2.0
----|------|------
2 |8.0 | 1.0
----|------|------
3 |5.0 | 1.0
----|------|------
4 |2.2 | 0.0
调用示例:
Scala:
Java:
Python:
算法介绍:
VectorIndexer解决数据集中的类别特征Vector。它可以自动识别哪些特征是类别型的,并且将原始值转换为类别指标。它的处理流程如下:
1.获得一个向量类型的输入以及maxCategories参数。
2.基于原始数值识别哪些特征需要被类别化,其中最多maxCategories需要被类别化。
3.对于每一个类别特征计算0-based类别指标。
4.对类别特征进行索引然后将原始值转换为指标。
索引后的类别特征可以帮助决策树等算法处理类别型特征,并得到较好结果。
在下面的例子中,我们读入一个数据集,然后使用VectorIndexer来决定哪些特征需要被作为非数值类型处理,将非数值型特征转换为他们的索引。
调用示例:
Scala:
import org.apache.spark.ml.feature.VectorIndexer
val data = spark.read.format("libsvm").load("data/mllib/sample_libsvm_data.txt")
val indexer = new VectorIndexer()
.setInputCol("features")
.setOutputCol("indexed")
.setMaxCategories(10)
val indexerModel = indexer.fit(data)
val categoricalFeatures: Set[Int] = indexerModel.categoryMaps.keys.toSet
println(s"Chose ${categoricalFeatures.size} categorical features: " +
categoricalFeatures.mkString(", "))
// Create new column "indexed" with categorical values transformed to indices
val indexedData = indexerModel.transform(data)
indexedData.show()Java:
import java.util.Map;
import org.apache.spark.ml.feature.VectorIndexer;
import org.apache.spark.ml.feature.VectorIndexerModel;
import org.apache.spark.sql.Dataset;
import org.apache.spark.sql.Row;
Dataset<Row> data = spark.read().format("libsvm").load("data/mllib/sample_libsvm_data.txt");
VectorIndexer indexer = new VectorIndexer()
.setInputCol("features")
.setOutputCol("indexed")
.setMaxCategories(10);
VectorIndexerModel indexerModel = indexer.fit(data);
Map<Integer, Map<Double, Integer>> categoryMaps = indexerModel.javaCategoryMaps();
System.out.print("Chose " + categoryMaps.size() + " categorical features:");
for (Integer feature : categoryMaps.keySet()) {
System.out.print(" " + feature);
}
System.out.println();
// Create new column "indexed" with categorical values transformed to indices
Dataset<Row> indexedData = indexerModel.transform(data);
indexedData.show();Python:
from pyspark.ml.feature import VectorIndexer
data = spark.read.format("libsvm").load("data/mllib/sample_libsvm_data.txt")
indexer = VectorIndexer(inputCol="features", outputCol="indexed", maxCategories=10)
indexerModel = indexer.fit(data)
# Create new column "indexed" with categorical values transformed to indices
indexedData = indexerModel.transform(data)
indexedData.show()Normalizer(正则化)
算法介绍:
Normalizer是一个转换器,它可以将多行向量输入转化为统一的形式。参数为p(默认值:2)来指定正则化中使用的p-norm。正则化操作可以使输入数据标准化并提高后期学习算法的效果。
下面的例子展示如何读入一个libsvm格式的数据,然后将每一行转换为
以及
形式。
调用示例:
Scala:
import org.apache.spark.ml.feature.Normalizer
val dataFrame = spark.read.format("libsvm").load("data/mllib/sample_libsvm_data.txt")
// Normalize each Vector using $L^1$ norm.
val normalizer = new Normalizer()
.setInputCol("features")
.setOutputCol("normFeatures")
.setP(1.0)
val l1NormData = normalizer.transform(dataFrame)
l1NormData.show()
// Normalize each Vector using $L^\infty$ norm.
val lInfNormData = normalizer.transform(dataFrame, normalizer.p -> Double.PositiveInfinity)
lInfNormData.show()Java:
import org.apache.spark.ml.feature.Normalizer;
import org.apache.spark.sql.Dataset;
import org.apache.spark.sql.Row;
Dataset<Row> dataFrame =
spark.read().format("libsvm").load("data/mllib/sample_libsvm_data.txt");
// Normalize each Vector using $L^1$ norm.
Normalizer normalizer = new Normalizer()
.setInputCol("features")
.setOutputCol("normFeatures")
.setP(1.0);
Dataset<Row> l1NormData = normalizer.transform(dataFrame);
l1NormData.show();
// Normalize each Vector using $L^\infty$ norm.
Dataset<Row> lInfNormData =
normalizer.transform(dataFrame, normalizer.p().w(Double.POSITIVE_INFINITY));
lInfNormData.show();Python:
from pyspark.ml.feature import Normalizer
dataFrame = spark.read.format("libsvm").load("data/mllib/sample_libsvm_data.txt")
# Normalize each Vector using $L^1$ norm.
normalizer = Normalizer(inputCol="features", outputCol="normFeatures", p=1.0)
l1NormData = normalizer.transform(dataFrame)
l1NormData.show()
# Normalize each Vector using $L^\infty$ norm.
lInfNormData = normalizer.transform(dataFrame, {normalizer.p: float("inf")})
lInfNormData.show()StandardScaler
算法介绍:
StandardScaler处理Vector数据,标准化每个特征使得其有统一的标准差以及(或者)均值为零。它需要如下参数:
1. withStd:默认值为真,使用统一标准差方式。
2. withMean:默认为假。此种方法将产出一个稠密输出,所以不适用于稀疏输入。
StandardScaler是一个Estimator,它可以fit数据集产生一个StandardScalerModel,用来计算汇总统计。然后产生的模可以用来转换向量至统一的标准差以及(或者)零均值特征。注意如果特征的标准差为零,则该特征在向量中返回的默认值为0.0。
下面的示例展示如果读入一个libsvm形式的数据以及返回有统一标准差的标准化特征。
调用示例:
Scala:
import org.apache.spark.ml.feature.StandardScaler
val dataFrame = spark.read.format("libsvm").load("data/mllib/sample_libsvm_data.txt")
val scaler = new StandardScaler()
.setInputCol("features")
.setOutputCol("scaledFeatures")
.setWithStd(true)
.setWithMean(false)
// Compute summary statistics by fitting the StandardScaler.
val scalerModel = scaler.fit(dataFrame)
// Normalize each feature to have unit standard deviation.
val scaledData = scalerModel.transform(dataFrame)
scaledData.show()Java:
import org.apache.spark.ml.feature.StandardScaler;
import org.apache.spark.ml.feature.StandardScalerModel;
import org.apache.spark.sql.Dataset;
import org.apache.spark.sql.Row;
Dataset<Row> dataFrame =
spark.read().format("libsvm").load("data/mllib/sample_libsvm_data.txt");
StandardScaler scaler = new StandardScaler()
.setInputCol("features")
.setOutputCol("scaledFeatures")
.setWithStd(true)
.setWithMean(false);
// Compute summary statistics by fitting the StandardScaler
StandardScalerModel scalerModel = scaler.fit(dataFrame);
// Normalize each feature to have unit standard deviation.
Dataset<Row> scaledData = scalerModel.transform(dataFrame);
scaledData.show();Python:
from pyspark.ml.feature import StandardScaler
dataFrame = spark.read.format("libsvm").load("data/mllib/sample_libsvm_data.txt")
scaler = StandardScaler(inputCol="features", outputCol="scaledFeatures",
withStd=True, withMean=False)
# Compute summary statistics by fitting the StandardScaler
scalerModel = scaler.fit(dataFrame)
# Normalize each feature to have unit standard deviation.
scaledData = scalerModel.transform(dataFrame)
scaledData.show()MinMaxScaler
算法介绍:
MinMaxScaler通过重新调节大小将Vector形式的列转换到指定的范围内,通常为[0,1],它的参数有:
1. min:默认为0.0,为转换后所有特征的下边界。
2. max:默认为1.0,为转换后所有特征的下边界。
MinMaxScaler计算数据集的汇总统计量,并产生一个MinMaxScalerModel。该模型可以将独立的特征的值转换到指定的范围内。
对于特征E来说,调整后的特征值如下:
如果
,则
。
注意因为零值转换后可能变为非零值,所以即便为稀疏输入,输出也可能为稠密向量。
下面的示例展示如果读入一个libsvm形式的数据以及调整其特征值到[0,1]之间。
调用示例:
Scala:
import org.apache.spark.ml.feature.MinMaxScaler
val dataFrame = spark.read.format("libsvm").load("data/mllib/sample_libsvm_data.txt")
val scaler = new MinMaxScaler()
.setInputCol("features")
.setOutputCol("scaledFeatures")
// Compute summary statistics and generate MinMaxScalerModel
val scalerModel = scaler.fit(dataFrame)
// rescale each feature to range [min, max].
val scaledData = scalerModel.transform(dataFrame)
scaledData.show()Java:
import org.apache.spark.ml.feature.MinMaxScaler;
import org.apache.spark.ml.feature.MinMaxScalerModel;
import org.apache.spark.sql.Dataset;
import org.apache.spark.sql.Row;
Dataset<Row> dataFrame = spark
.read()
.format("libsvm")
.load("data/mllib/sample_libsvm_data.txt");
MinMaxScaler scaler = new MinMaxScaler()
.setInputCol("features")
.setOutputCol("scaledFeatures");
// Compute summary statistics and generate MinMaxScalerModel
MinMaxScalerModel scalerModel = scaler.fit(dataFrame);
// rescale each feature to range [min, max].
Dataset<Row> scaledData = scalerModel.transform(dataFrame);
scaledData.show();Python:
from pyspark.ml.feature import MinMaxScaler
dataFrame = spark.read.format("libsvm").load("data/mllib/sample_libsvm_data.txt")
scaler = MinMaxScaler(inputCol="features", outputCol="scaledFeatures")
# Compute summary statistics and generate MinMaxScalerModel
scalerModel = scaler.fit(dataFrame)
# rescale each feature to range [min, max].
scaledData = scalerModel.transform(dataFrame)
scaledData.show()MaxAbsScaler
算法介绍:
MaxAbsScaler使用每个特征的最大值的绝对值将输入向量的特征值转换到[-1,1]之间。因为它不会转移/集中数据,所以不会破坏数据的稀疏性。
下面的示例展示如果读入一个libsvm形式的数据以及调整其特征值到[-1,1]之间。
调用示例:
Scala:
import org.apache.spark.ml.feature.MaxAbsScaler
val dataFrame = spark.read.format("libsvm").load("data/mllib/sample_libsvm_data.txt")
val scaler = new MaxAbsScaler()
.setInputCol("features")
.setOutputCol("scaledFeatures")
// Compute summary statistics and generate MaxAbsScalerModel
val scalerModel = scaler.fit(dataFrame)
// rescale each feature to range [-1, 1]
val scaledData = scalerModel.transform(dataFrame)
scaledData.show()Java:
import org.apache.spark.ml.feature.MaxAbsScaler;
import org.apache.spark.ml.feature.MaxAbsScalerModel;
import org.apache.spark.sql.Dataset;
import org.apache.spark.sql.Row;
Dataset<Row> dataFrame = spark
.read()
.format("libsvm")
.load("data/mllib/sample_libsvm_data.txt");
MaxAbsScaler scaler = new MaxAbsScaler()
.setInputCol("features")
.setOutputCol("scaledFeatures");
// Compute summary statistics and generate MaxAbsScalerModel
MaxAbsScalerModel scalerModel = scaler.fit(dataFrame);
// rescale each feature to range [-1, 1].
Dataset<Row> scaledData = scalerModel.transform(dataFrame);
scaledData.show();Python:
from pyspark.ml.feature import MaxAbsScaler
dataFrame = spark.read.format("libsvm").load("data/mllib/sample_libsvm_data.txt")
scaler = MaxAbsScaler(inputCol="features", outputCol="scaledFeatures")
# Compute summary statistics and generate MaxAbsScalerModel
scalerModel = scaler.fit(dataFrame)
# rescale each feature to range [-1, 1].
scaledData = scalerModel.transform(dataFrame)
scaledData.show()Bucketizer
算法介绍:
Bucketizer将一列连续的特征转换为特征区间,区间由用户指定。参数如下:
1. splits:分裂数为n+1时,将产生n个区间。除了最后一个区间外,每个区间范围[x,y]由分裂的x,y决定。分裂必须是严格递增的。在分裂指定外的值将被归为错误。两个分裂的例子为Array(Double.NegativeInfinity,0.0,
1.0, Double.PositiveInfinity)以及Array(0.0, 1.0, 2.0)。
注意,当不确定分裂的上下边界时,应当添加Double.NegativeInfinity和Double.PositiveInfinity以免越界。
下面将展示Bucketizer的使用方法。
调用示例:
Scala:
import org.apache.spark.ml.feature.Bucketizer
val splits = Array(Double.NegativeInfinity, -0.5, 0.0, 0.5, Double.PositiveInfinity)
val data = Array(-0.5, -0.3, 0.0, 0.2)
val dataFrame = spark.createDataFrame(data.map(Tuple1.apply)).toDF("features")
val bucketizer = new Bucketizer()
.setInputCol("features")
.setOutputCol("bucketedFeatures")
.setSplits(splits)
// Transform original data into its bucket index.
val bucketedData = bucketizer.transform(dataFrame)
bucketedData.show()Java:
import java.util.List;
import org.apache.spark.ml.feature.Bucketizer;
import org.apache.spark.sql.Dataset;
import org.apache.spark.sql.Row;
import org.apache.spark.sql.RowFactory;
import org.apache.spark.sql.types.DataTypes;
import org.apache.spark.sql.types.Metadata;
import org.apache.spark.sql.types.StructField;
import org.apache.spark.sql.types.StructType;
double[] splits = {Double.NEGATIVE_INFINITY, -0.5, 0.0, 0.5, Double.POSITIVE_INFINITY};
List<Row> data = Arrays.asList(
RowFactory.create(-0.5),
RowFactory.create(-0.3),
RowFactory.create(0.0),
RowFactory.create(0.2)
);
StructType schema = new StructType(new StructField[]{
new StructField("features", DataTypes.DoubleType, false, Metadata.empty())
});
Dataset<Row> dataFrame = spark.createDataFrame(data, schema);
Bucketizer bucketizer = new Bucketizer()
.setInputCol("features")
.setOutputCol("bucketedFeatures")
.setSplits(splits);
// Transform original data into its bucket index.
Dataset<Row> bucketedData = bucketizer.transform(dataFrame);
bucketedData.show();Python:
from pyspark.ml.feature import Bucketizer
splits = [-float("inf"), -0.5, 0.0, 0.5, float("inf")]
data = [(-0.5,), (-0.3,), (0.0,), (0.2,)]
dataFrame = spark.createDataFrame(data, ["features"])
bucketizer = Bucketizer(splits=splits, inputCol="features", outputCol="bucketedFeatures")
# Transform original data into its bucket index.
bucketedData = bucketizer.transform(dataFrame)
bucketedData.show()ElementwiseProduct
算法介绍:
ElementwiseProduct按提供的“weight”向量,返回与输入向量元素级别的乘积。即是说,按提供的权重分别对输入数据进行缩放,得到输入向量v以及权重向量w的Hadamard积。
下面例子展示如何通过转换向量的值来调整向量。
调用示例:
Scala:
import org.apache.spark.ml.feature.ElementwiseProduct
import org.apache.spark.ml.linalg.Vectors
// Create some vector data; also works for sparse vectors
val dataFrame = spark.createDataFrame(Seq(
("a", Vectors.dense(1.0, 2.0, 3.0)),
("b", Vectors.dense(4.0, 5.0, 6.0)))).toDF("id", "vector")
val transformingVector = Vectors.dense(0.0, 1.0, 2.0)
val transformer = new ElementwiseProduct()
.setScalingVec(transformingVector)
.setInputCol("vector")
.setOutputCol("transformedVector")
// Batch transform the vectors to create new column:
transformer.transform(dataFrame).show()Java:
import java.util.ArrayList;
import java.util.Arrays;
import java.util.List;
import org.apache.spark.ml.feature.ElementwiseProduct;
import org.apache.spark.ml.linalg.Vector;
import org.apache.spark.ml.linalg.VectorUDT;
import org.apache.spark.ml.linalg.Vectors;
import org.apache.spark.sql.Row;
import org.apache.spark.sql.RowFactory;
import org.apache.spark.sql.types.DataTypes;
import org.apache.spark.sql.types.StructField;
import org.apache.spark.sql.types.StructType;
// Create some vector data; also works for sparse vectors
List<Row> data = Arrays.asList(
RowFactory.create("a", Vectors.dense(1.0, 2.0, 3.0)),
RowFactory.create("b", Vectors.dense(4.0, 5.0, 6.0))
);
List<StructField> fields = new ArrayList<>(2);
fields.add(DataTypes.createStructField("id", DataTypes.StringType, false));
fields.add(DataTypes.createStructField("vector", new VectorUDT(), false));
StructType schema = DataTypes.createStructType(fields);
Dataset<Row> dataFrame = spark.createDataFrame(data, schema);
Vector transformingVector = Vectors.dense(0.0, 1.0, 2.0);
ElementwiseProduct transformer = new ElementwiseProduct()
.setScalingVec(transformingVector)
.setInputCol("vector")
.setOutputCol("transformedVector");
// Batch transform the vectors to create new column:
transformer.transform(dataFrame).show();Python:
from pyspark.ml.feature import ElementwiseProduct from pyspark.ml.linalg import Vectors # Create some vector data; also works for sparse vectors data = [(Vectors.dense([1.0, 2.0, 3.0]),), (Vectors.dense([4.0, 5.0, 6.0]),)] df = spark.createDataFrame(data, ["vector"]) transformer = ElementwiseProduct(scalingVec=Vectors.dense([0.0, 1.0, 2.0]), inputCol="vector", outputCol="transformedVector") # Batch transform the vectors to create new column: transformer.transform(df).show()
SQLTransformer
算法介绍:
SQLTransformer工具用来转换由SQL定义的陈述。目前仅支持SQL语法如"SELECT
...FROM __THIS__ ...",其中"__THIS__"代表输入数据的基础表。选择语句指定输出中展示的字段、元素和表达式,支持Spark SQL中的所有选择语句。用户可以基于选择结果使用Spark
SQL建立方程或者用户自定义函数。SQLTransformer支持语法示例如下:
1. SELECTa, a + b AS a_b FROM __THIS__
2. SELECTa, SQRT(b) AS b_sqrt FROM __THIS__ where a > 5
3. SELECTa, b, SUM(c) AS c_sum FROM __THIS__ GROUP BY a, b
示例:
假设我们有如下DataFrame包含id,v1,v2列:
id | v1 | v2
----|-----|-----
0 | 1.0 | 3.0
2 | 2.0 | 5.0
使用SQLTransformer语句"SELECT *,(v1 + v2) AS v3, (v1 * v2) AS v4 FROM __THIS__"转换后得到输出如下:
id | v1 | v2 | v3 | v4
----|-----|-----|-----|-----
0 | 1.0| 3.0 | 4.0 | 3.0
2 | 2.0| 5.0 | 7.0 |10.0
调用示例:
Scala:
import org.apache.spark.ml.feature.SQLTransformer
val df = spark.createDataFrame(
Seq((0, 1.0, 3.0), (2, 2.0, 5.0))).toDF("id", "v1", "v2")
val sqlTrans = new SQLTransformer().setStatement(
"SELECT *, (v1 + v2) AS v3, (v1 * v2) AS v4 FROM __THIS__")
sqlTrans.transform(df).show()Java:
import java.util.Arrays;
import java.util.List;
import org.apache.spark.ml.feature.SQLTransformer;
import org.apache.spark.sql.Dataset;
import org.apache.spark.sql.Row;
import org.apache.spark.sql.RowFactory;
import org.apache.spark.sql.SparkSession;
import org.apache.spark.sql.types.*;
List<Row> data = Arrays.asList(
RowFactory.create(0, 1.0, 3.0),
RowFactory.create(2, 2.0, 5.0)
);
StructType schema = new StructType(new StructField [] {
new StructField("id", DataTypes.IntegerType, false, Metadata.empty()),
new StructField("v1", DataTypes.DoubleType, false, Metadata.empty()),
new StructField("v2", DataTypes.DoubleType, false, Metadata.empty())
});
Dataset<Row> df = spark.createDataFrame(data, schema);
SQLTransformer sqlTrans = new SQLTransformer().setStatement(
"SELECT *, (v1 + v2) AS v3, (v1 * v2) AS v4 FROM __THIS__");
sqlTrans.transform(df).show();Python:
from pyspark.ml.feature import SQLTransformer df = spark.createDataFrame([ (0, 1.0, 3.0), (2, 2.0, 5.0) ], ["id", "v1", "v2"]) sqlTrans = SQLTransformer( statement="SELECT *, (v1 + v2) AS v3, (v1 * v2) AS v4 FROM __THIS__") sqlTrans.transform(df).show()
VectorAssembler
算法介绍:
VectorAssembler是一个转换器,它将给定的若干列合并为一列向量。它可以将原始特征和一系列通过其他转换器得到的特征合并为单一的特征向量,来训练如逻辑回归和决策树等机器学习算法。VectorAssembler可接受的输入列类型:数值型、布尔型、向量型。输入列的值将按指定顺序依次添加到一个新向量中。
示例:
假设我们有如下DataFrame包含id,hour,mobile,
userFeatures以及clicked列:
id | hour | mobile| userFeatures | clicked
----|------|--------|------------------|---------
0 |18 | 1.0 | [0.0, 10.0, 0.5] | 1.0
userFeatures列中含有3个用户特征。我们想将hour,mobile以及userFeatures合并为一个新列。将VectorAssembler的输入指定为hour,mobile以及userFeatures,输出指定为features,通过转换我们将得到以下结果:
id | hour | mobile| userFeatures | clicked | features
----|------|--------|------------------|---------|-----------------------------
0 |18 | 1.0 | [0.0, 10.0, 0.5] | 1.0 | [18.0, 1.0, 0.0, 10.0, 0.5]
调用示例:
Scala:
import org.apache.spark.ml.feature.VectorAssembler
import org.apache.spark.ml.linalg.Vectors
val dataset = spark.createDataFrame(
Seq((0, 18, 1.0, Vectors.dense(0.0, 10.0, 0.5), 1.0))
).toDF("id", "hour", "mobile", "userFeatures", "clicked")
val assembler = new VectorAssembler()
.setInputCols(Array("hour", "mobile", "userFeatures"))
.setOutputCol("features")
val output = assembler.transform(dataset)
println(output.select("features", "clicked").first())Java:
import java.util.Arrays;
import org.apache.spark.ml.feature.VectorAssembler;
import org.apache.spark.ml.linalg.VectorUDT;
import org.apache.spark.ml.linalg.Vectors;
import org.apache.spark.sql.Dataset;
import org.apache.spark.sql.Row;
import org.apache.spark.sql.RowFactory;
import org.apache.spark.sql.types.*;
import static org.apache.spark.sql.types.DataTypes.*;
StructType schema = createStructType(new StructField[]{
createStructField("id", IntegerType, false),
createStructField("hour", IntegerType, false),
createStructField("mobile", DoubleType, false),
createStructField("userFeatures", new VectorUDT(), false),
createStructField("clicked", DoubleType, false)
});
Row row = RowFactory.create(0, 18, 1.0, Vectors.dense(0.0, 10.0, 0.5), 1.0);
Dataset<Row> dataset = spark.createDataFrame(Arrays.asList(row), schema);
VectorAssembler assembler = new VectorAssembler()
.setInputCols(new String[]{"hour", "mobile", "userFeatures"})
.setOutputCol("features");
Dataset<Row> output = assembler.transform(dataset);
System.out.println(output.select("features", "clicked").first());Python:
from pyspark.ml.linalg import Vectors
from pyspark.ml.feature import VectorAssembler
dataset = spark.createDataFrame(
[(0, 18, 1.0, Vectors.dense([0.0, 10.0, 0.5]), 1.0)],
["id", "hour", "mobile", "userFeatures", "clicked"])
assembler = VectorAssembler(
inputCols=["hour", "mobile", "userFeatures"],
outputCol="features")
output = assembler.transform(dataset)
print(output.select("features", "clicked").first())QuantileDiscretizer
算法介绍:
QuantileDiscretizer讲连续型特征转换为分级类别特征。分级的数量由numBuckets参数决定。分级的范围有渐进算法决定。渐进的精度由relativeError参数决定。当relativeError设置为0时,将会计算精确的分位点(计算代价较高)。分级的上下边界为负无穷到正无穷,覆盖所有的实数值。
示例:
假设我们有如下DataFrame包含id,hour:
id | hour
----|------
0 |18.0
----|------
1 |19.0
----|------
2 | 8.0
----|------
3 | 5.0
----|------
4 | 2.2
hour是一个Double类型的连续特征,将参数numBuckets设置为3,我们可以将hour转换为如下分级特征。
id | hour | result
----|------|------
0 |18.0 | 2.0
----|------|------
1 |19.0 | 2.0
----|------|------
2 |8.0 | 1.0
----|------|------
3 |5.0 | 1.0
----|------|------
4 |2.2 | 0.0
调用示例:
Scala:
import org.apache.spark.ml.feature.QuantileDiscretizer
val data = Array((0, 18.0), (1, 19.0), (2, 8.0), (3, 5.0), (4, 2.2))
var df = spark.createDataFrame(data).toDF("id", "hour")
val discretizer = new QuantileDiscretizer()
.setInputCol("hour")
.setOutputCol("result")
.setNumBuckets(3)
val result = discretizer.fit(df).transform(df)
result.show()Java:
import java.util.Arrays;
import java.util.List;
import org.apache.spark.ml.feature.QuantileDiscretizer;
import org.apache.spark.sql.Dataset;
import org.apache.spark.sql.Row;
import org.apache.spark.sql.RowFactory;
import org.apache.spark.sql.types.DataTypes;
import org.apache.spark.sql.types.Metadata;
import org.apache.spark.sql.types.StructField;
import org.apache.spark.sql.types.StructType;
List<Row> data = Arrays.asList(
RowFactory.create(0, 18.0),
RowFactory.create(1, 19.0),
RowFactory.create(2, 8.0),
RowFactory.create(3, 5.0),
RowFactory.create(4, 2.2)
);
StructType schema = new StructType(new StructField[]{
new StructField("id", DataTypes.IntegerType, false, Metadata.empty()),
new StructField("hour", DataTypes.DoubleType, false, Metadata.empty())
});
Dataset<Row> df = spark.createDataFrame(data, schema);
QuantileDiscretizer discretizer = new QuantileDiscretizer()
.setInputCol("hour")
.setOutputCol("result")
.setNumBuckets(3);
Dataset<Row> result = discretizer.fit(df).transform(df);
result.show();Python:
from pyspark.ml.feature import QuantileDiscretizer data = [(0, 18.0,), (1, 19.0,), (2, 8.0,), (3, 5.0,), (4, 2.2,)] df = spark.createDataFrame(data, ["id", "hour"]) discretizer = QuantileDiscretizer(numBuckets=3, inputCol="hour", outputCol="result") result = discretizer.fit(df).transform(df) result.show()
相关文章推荐
- 二十种特征变换方法及Spark MLlib调用实例(Scala/Java/python)(一)
- spark--二十种特征变换方法及Spark MLlib调用实例(Scala/Java/python)(一)
- 二十种特征变换方法及Spark MLlib调用实例(Scala/Java/python)(二)
- 二十种特征变换方法及Spark MLlib调用实例(Scala/Java/python)(二)
- 二十种特征变换方法及Spark MLlib调用实例(Scala/Java/python)(一)
- 二十种特征变换方法及Spark MLlib调用实例(Scala/Java/python)(二)
- 三种特征选择方法及Spark MLlib调用实例(Scala/Java/python)
- 三种特征选择方法及Spark MLlib调用实例(Scala/Java/python)
- 三种文本特征提取(TF-IDF/Word2Vec/CountVectorizer)及Spark MLlib调用实例(Scala/Java/python)
- scala--三种文本特征提取(TF-IDF/Word2Vec/CountVectorizer)及Spark MLlib调用实例(Scala/Java/python)
- 逻辑回归算法原理及Spark MLlib调用实例(Scala/Java/python)
- 决策树算法原理及Spark MLlib调用实例(Scala/Java/python)
- 随机森林(Random Forest)算法原理及Spark MLlib调用实例(Scala/Java/python)
- 梯度迭代树(GBDT)算法原理及Spark MLlib调用实例(Scala/Java/python)
- 多层感知机(MLP)算法原理及Spark MLlib调用实例(Scala/Java/Python)
- One-vs-Rest算法介绍及Spark MLlib调用实例(Scala/Java/Python)
- 朴素贝叶斯算法原理及Spark MLlib调用实例(Scala/Java/Python)
- 广义线性模型(GLMs)算法原理及Spark MLlib调用实例(Scala/Java/Python)
- 决策树回归算法原理及Spark MLlib调用实例(Scala/Java/python)
- 随机森林回归(Random Forest)算法原理及Spark MLlib调用实例(Scala/Java/python)
