《机器学习》周志华 习题答案9.4
2016-07-06 22:12
387 查看
原题采用Kmeans方法对西瓜数据集进行聚类。我花了一些时间居然没找到西瓜数据集4.0在哪里,于是直接采用sklearn给的例子来分析一遍,更能说明Kmeans的效果。
运行文本结果:
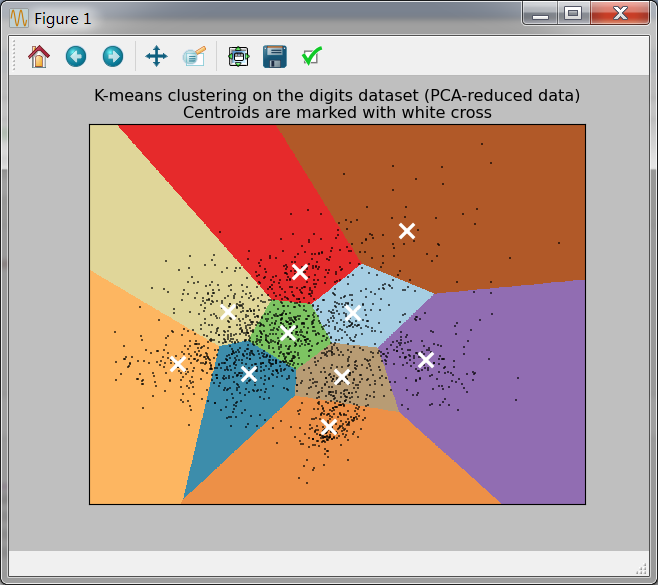
图片结果:
#!/usr/bin/python
# -*- coding:utf-8 -*-
import numpy as np
import matplotlib.pyplot as plt
from sklearn.ensemble import BaggingClassifier
from sklearn.tree import DecisionTreeClassifier
file1 = open('c:\quant\watermelon.csv','r')
data = [line.strip('\n').split(',') for line in file1]
data = np.array(data)
#X = [[float(raw[-7]),float(raw[-6]),float(raw[-5]),float(raw[-4]),float(raw[-3]), float(raw[-2])] for raw in data[1:,1:-1]]
X = [[float(raw[-3]), float(raw[-2])] for raw in data[1:]]
y = [1 if raw[-1]=='1' else 0 for raw in data[1:]]
X = np.array(X)
y = np.array(y)
print(__doc__)
from time import time
import numpy as np
import matplotlib.pyplot as plt
from sklearn import metrics
from sklearn.cluster import KMeans
from sklearn.datasets import load_digits
from sklearn.decomposition import PCA
from sklearn.preprocessing import scale
np.random.seed(42)
digits = load_digits()
data = scale(digits.data)
n_samples, n_features = data.shape
n_digits = len(np.unique(digits.target))
labels = digits.target
sample_size = 300
print("n_digits: %d, \t n_samples %d, \t n_features %d"
% (n_digits, n_samples, n_features))
#一共十个不同的类
print(79 * '_')
print('% 9s' % 'init'
' time inertia homo compl v-meas ARI AMI silhouette')
def bench_k_means(estimator, name, data):
t0 = time()
estimator.fit(data)
print('% 9s %.2fs %i %.3f %.3f %.3f %.3f %.3f %.3f'
% (name, (time() - t0), estimator.inertia_,
metrics.homogeneity_score(labels, estimator.labels_),
metrics.completeness_score(labels, estimator.labels_),
metrics.v_measure_score(labels, estimator.labels_),
metrics.adjusted_rand_score(labels, estimator.labels_),
metrics.adjusted_mutual_info_score(labels, estimator.labels_),
metrics.silhouette_score(data, estimator.labels_,
metric='euclidean',
sample_size=sample_size)))
#Homogeneity 和 completeness 表示簇的均一性和完整性。V值是他们的调和平均,值越大,说明效果越好。
bench_k_means(KMeans(init='k-means++', n_clusters=n_digits, n_init=10),
name="k-means++", data=data)
bench_k_means(KMeans(init='random', n_clusters=n_digits, n_init=10),
name="random", data=data)
# in this case the seeding of the centers is deterministic, hence we run the
# kmeans algorithm only once with n_init=1
pca = PCA(n_components=n_digits).fit(data)
bench_k_means(KMeans(init=pca.components_, n_clusters=n_digits, n_init=1),
name="PCA-based",
data=data)
print(79 * '_')
###############################################################################
# Visualize the results on PCA-reduced data
reduced_data = PCA(n_components=2).fit_transform(data)
kmeans = KMeans(init='k-means++', n_clusters=n_digits, n_init=10)
kmeans.fit(reduced_data)
# Step size of the mesh. Decrease to increase the quality of the VQ.
h = .02 # point in the mesh [x_min, m_max]x[y_min, y_max].
# Plot the decision boundary. For that, we will assign a color to each
x_min, x_max = reduced_data[:, 0].min() - 1, reduced_data[:, 0].max() + 1
y_min, y_max = reduced_data[:, 1].min() - 1, reduced_data[:, 1].max() + 1
xx, yy = np.meshgrid(np.arange(x_min, x_max, h), np.arange(y_min, y_max, h))
# Obtain labels for each point in mesh. Use last trained model.
Z = kmeans.predict(np.c_[xx.ravel(), yy.ravel()])
# Put the result into a color plot
Z = Z.reshape(xx.shape)
plt.figure(1)
plt.clf()
plt.imshow(Z, interpolation='nearest',
extent=(xx.min(), xx.max(), yy.min(), yy.max()),
cmap=plt.cm.Paired,
aspect='auto', origin='lower')
plt.plot(reduced_data[:, 0], reduced_data[:, 1], 'k.', markersize=2)
# Plot the centroids as a white X
centroids = kmeans.cluster_centers_
plt.scatter(centroids[:, 0], centroids[:, 1],
marker='x', s=169, linewidths=3,
color='w', zorder=10)
plt.title('K-means clustering on the digits dataset (PCA-reduced data)\n'
'Centroids are marked with white cross')
plt.xlim(x_min, x_max)
plt.ylim(y_min, y_max)
plt.xticks(())
plt.yticks(())
plt.show()运行文本结果:
n_digits: 10, n_samples 1797, n_features 64 _______________________________________________________________________________ init time inertia homo compl v-meas ARI AMI silhouette k-means++ 0.21s 69432 0.602 0.650 0.625 0.465 0.598 0.146 random 0.20s 69694 0.669 0.710 0.689 0.553 0.666 0.147 PCA-based 0.02s 71820 0.673 0.715 0.693 0.567 0.670 0.150 我们可以看到降维处理后运行时间缩短,而且V值还略高于以上两种方法。
图片结果:

相关文章推荐
- linux执行sh脚本文件命令
- 顺序容器的功能对比
- 使用console调试JavaScript(一)
- iOS可变参数函数的编写
- ADS下的分散加载文件应用实例
- linux下的C语言开发(gdb调试)
- 【Arduino官方教程】数字处理示例(三):按键防抖
- logstash实现日志文件同步到elasticsearch深入详解
- 盒子嵌套
- linux下安装ffmpeg
- KindEditor编辑器关闭过滤html,js,css标题方法
- POJ 3784 Running Median
- ARM 必须知道的知识
- MyEclipse破解失败的解决办法 ---谷营中西
- JVM内存区域
- Python & virtualenv使用说明
- 7月5日实习日志
- Java 身份证号码验证程序
- C++多线程编程简单实例
- Java中线程通信协作
