吴恩达 深度学习第三周 浅层神经网络 logistic_regression python代码实现
2017-12-07 09:49
1116 查看
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
#%matplotlib inline
from sklearn.datasets import load_iris
from sklearn.model_selection import train_test_split
iris = load_iris()
x = iris.data
y = iris.target
for i in range(len(y)):
if y[i] == 2: y[i] =1
#print(y)
x_train,x_test, y_train, y_test = train_test_split(x,y,test_size = 0.3)
x_train = x_train.T
x_test = x_test.T
y_train = y_train.reshape(1,-1)
y_tset = y_test.reshape(1,-1)
print(y_test)
num_n1 = 4
num_n2 = 1
num_n0 = x_train.shape[0]
m_sample = x_train.shape[1]
learning_rate = 0.005
w1 = np.random.randn(num_n1, num_n0)*0.01
b1 = np.random.randn(num_n1).reshape(-1,1)*0.01
w2 = np.random.randn(num_n2,num_n1)*0.01
b2 = np.random.randn(num_n2).reshape(-1,1)*0.01
parameters =dict(w1=w1,w2=w2,b1=b1,b2=b2)
print(parameters['w2'],parameters['b2'])
def forward(x,parameters):
#set parameters
w1 = parameters['w1'] # shape n1 * n0
w2 = parameters['w2'] # shape n2 * n1
b1 = parameters['b1'] # shape n1 * 1
b2 = parameters['b2'] # shape n2 * 1 actuall n2 = 1
# cacl internal matrix
Z1 = np.dot(w1,x)+b1 # shape n1 * m
A1 = 1.0/(1.0+np.exp(-Z1)) # shape n1 * m
Z2 = np.dot(w2,A1) + b2 # shape n2 * m
A2 = 1.0/(1.0 + np.exp(-Z2)) # shape n2 * m
# matrix shape num
m = x.shape[1]
n0 = x.shape[0]
n1 = w1.shape[0]
n2 = w2.shape[0]
#print('correct param m,n0,n1,n2\n',m,n0,n1,n2)
assert(w2.shape[1] == n1)
assert(Z1.shape[0] == n1 )
assert( Z1.shape[1]==m)
assert(A1.shape[0]== n1 )
assert(A1.shape[1]==m)
assert(Z2.shape[0]==n2 )
assert(Z2.shape[1]==m)
assert(A2.shape[0]==n2 )
assert(A2.shape[1]==m)
# return dict of internal matrix
temp = dict(Z1 = Z1,
A1=A1,
Z2=Z2,
A2=A2)
return temp
# backpropagation function
def backward(X,Y,temp,parameter):
'''
INPUTS:
X: input training feature datas shape n0 * m
Y: input training labeled datas shape 1 * m
temp: dict of transient matrix A1,Z1,A2,Z2
parameter: dict structure , includeing : w1,b1,w2,b2
OUTPUS:
dZ2: used calc deriative
dw2: gradient shape of
db2: gradient shape of
dZ1: gradient shape of
dw1: gradient shape of
db1: gradient shape of
'''
# setting of internal matrix values
A2 = temp['A2'] #shape n2 * m
A1 = temp['A1'] #shape n1 * m
Z1 = temp['Z1'] #shape n1 * m
Z2 = temp['Z2'] #shape n2 * m
w2 = parameters['w2'] # shape n2 * n1
m_sample = X.shape[1]
# calc gradient values in layer2
dZ2 = A2 - Y # shape n2 * m
dw2 = np.dot(dZ2, A1.T) # shape n2 * n1
dw2 = dw2/np.float(m_sample)
db2 = np.sum(dZ2,axis=1,keepdims=True) # shape n2 * 1
db2 = db2/np.float(m_sample)
# calc gridient values in layer1
GP = np.multiply(A1,(1.0-A1)) # shape n1 * m
dZ1 = np.multiply(np.dot(w2.T,dZ2),GP) # shape n1 * m
dw1 = np.dot(dZ1,X.T) # shape n1 * n0
dw1 = dw1/np.float(m_sample)
db1 = np.sum(dZ1,axis=1,keepdims=True) # shape n1 * 1
db1 = db1/np.float(m_sample)
#----------------
#-- check dims---
#----------------
n0 = X.shape[0]
m = X.shape[1]
n1 = A1.shape[0]
n2 = A2.shape[0]
assert(dZ2.shape == Z2.shape)
assert(dw2.shape == w2.shape)
assert(db2.shape[0] == n2)
assert(dZ1.shape == Z1.shape)
assert(dw1.shape[0]==n1)
assert(dw1.shape[1]==n0)
assert(db1.shape[0]==n1)
assert(GP.shape == A1.shape)
# set return values of gradients in two layers
temp = dict(dZ2 = dZ2,
dw2 = dw2,
db2 = db2,
dZ1 = dZ1,
dw1 = dw1,
db1 = db1)
return temp
# loss function
def get_loss(Y,catch):
'''
INPUTS:
Y: input values shape 1 * m
catch : temp dict Z2,A2,Z1,A1 values
A2: matrix used to calc loss shape : (n2 * m actually 1 * m)
OUTPUS:
return loss values
loss: return values shape: real number
'''
m = Y.shape[1]
A2 = catch['A2']
loss = np.dot(Y,(np.log(A2)).T) + np.dot((1.0-Y),np.log(1.0-A2).T)
loss = -loss/np.float(m)
return loss
# prediction function used to predict accuracy
def predict(X,Y,parameters):
'''
function used to predict the results
INPUTS:
X: input matrix shape: n0 * m
Y: input matrix shape: n2 * m actually 1 * m
parameters: w1,b1,w2,b2
OUTPUS :
accuracy : score in classification (real number)
'''
catch = forward(X,parameters)
A2 = catch['A2']
A2 = A2.reshape(-1,1)
Y = Y.reshape(-1,1)
error_count = 0
print(A2.shape,Y.shape)
assert(len(Y) == len(A2))
for i in range(len(A2)):
if A2[i] >= 0.5:
A2[i] = 1
else:
A2[i] = 0
for i in range(len(Y)):
if Y[i] != A2[i]:
error_count +=1
accuracy =1.0 - np.float(error_count)/np.float(len(Y))
print('The Accuracy of network is:', accuracy)
return accuracy
# function used to learning parameters
def parameter_learning(X,Y,X_test,Y_test,parameters,learning_rate):
'''
INPUTS:
X: input values feats data shape n0 * m
Y: input values of labeled data shape 1 * m
parameters: including w1,b1, w2,b2 using to describe models
w1: first layer weighted values shape: n1 * n0
b1: first layer concate shape: n1 * 1
w2: second layer weighted values shape : n2 * n1
b2: second layer concate shape: n2 * 1
learning_rate: learning_rate
OUTPUTS:
parameter: final convergenced parameters
dloss: the relative errors of last two iterations
'''
epoch_num = 10000
w1 = parameters['w1'] # 此处的赋值,仅仅是把地址交给了变量w1 改变w1 其对应的 parameters中w1 也改变
w2 = parameters['w2']
b1 = parameters['b1']
b2 = parameters['b2']
plt.ion()
train_accuracy= []
test_accuracy = []
steps = []
residual = []
plt.figure(1,figsize=(6,6))
plt.figure(2,figsize=(6,6))
for i in range(epoch_num):
catch = forward(X,parameters)
gradient = backward(X,Y,catch, parameters)
loss = get_loss(Y,catch)
loss_temp = loss
#gradient
dw1 = gradient['dw1']
db1 = gradient['db1']
dw2 = gradient['dw2']
db2 = gradient['db2']
# update parameter values
w1 += -learning_rate * dw1
b1 += -learning_rate * db1
w2 += -learning_rate * dw2
b2 += -learning_rate * db2
catch = forward(X,parameters)
loss = get_loss(Y,catch)
dloss = np.abs(loss-loss_temp)/(np.abs(loss)+0.0000001)
# set of output and figures
if(i%100 == 0):
print('iteration num = ',i, 'dloss =', dloss,loss)
steps.append(i)
residual.append(dloss[0])
plt.figure(1)
train_accuracy.append(predict(X,Y,parameters))
test_accuracy.append(predict(X_test,Y_test,parameters))
line1,=plt.plot(steps,train_accuracy,'red',linewidth=1.5,label='Train Accuracy')
line2,=plt.plot(steps,test_accuracy, 'green', linewidth=1.5,label='Test Accuracy')
plt.ylim([0,1.3])
plt.xlabel('Trainning steps')
plt.ylabel('Trainning accuracy')
plt.title('train/test VS steps')
plt.legend([line1,line2],['Train Accuracy','Test Accuracy'], loc='upper right')
plt.figure(2)
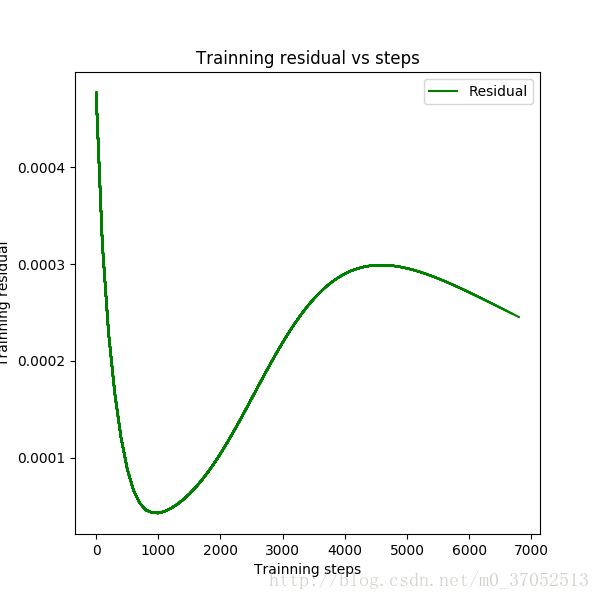
line3, = plt.plot(steps,residual,'g-',linewidth=1.5,markersize=4,label='residual')
plt.xlabel('Trainning steps')
plt.ylabel('Trainning residual')
plt.title('Trainning residual vs steps')
plt.legend([line3],['Residual'],loc='upper right')
plt.pause(0.01)
# define convergence criterion
if dloss <=1.0e-6:
print('iteration convergenced!!',dloss,i)
return parameters,dloss
return parameters,dloss
final_params,dloss = parameter_learning(x_train,y_train,x_test,y_test,parameters,learning_rate)
print('final loss:',dloss)
#print(final_params)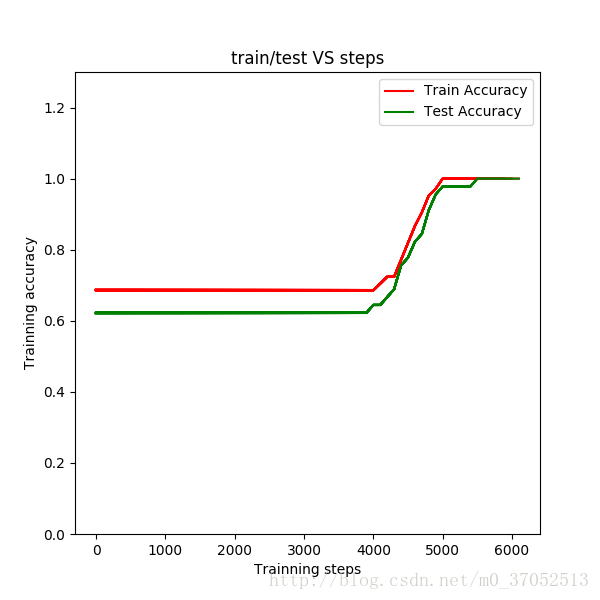
相关文章推荐
- 深度学习与神经网络-吴恩达(Part1Week4)-深度神经网络编程实现(python)-基础篇
- 深度学习论文-神经网络的代码实现(python版本)
- 深度学习与神经网络-吴恩达(Part1Week3)-单隐层神经网络编程实现(python)
- Coursera deep learning 吴恩达 神经网络和深度学习 第二周 编程作业 Logistic Regression with a Neural Network mindset
- coursera 吴恩达 -- 第一课 神经网络和深度学习 :第二周课后习题 Logistic Regression with a Neural Network mindset
- 吴恩达深度学习第一课第三周(浅层神经网络)
- [深度学习]Python/Theano实现逻辑回归网络的代码分析
- Coursera 深度学习 deep learning.ai 吴恩达 神经网络和深度学习 第一课 第二周 编程作业 Python Basics with Numpy
- 吴恩达老师深度学习视频课笔记:单隐含层神经网络公式推导及C++实现(二分类)
- 深度学习5:python实现三层神经网络
- Python实现深度学习之-神经网络识别手写数字(更新中,更新日期:2017-07-12)
- Python20行代码实现多层神经网络的学习
- Coursera deeplearning.ai 深度学习笔记1-3-Shallow Neural Networks-浅层神经网络原理推导与代码实现
- coursera 吴恩达 -- 第一课 神经网络和深度学习 :第三周课后习题 Shallow Neural Networks Quiz, 10 questions
- 神经网络与深度学习笔记(二)python 实现随机梯度下降
- Coursera deeplearning.ai 深度学习笔记2-1-Practical aspects of deep learning-神经网络实际问题分析(初始化&正则化&训练效率)与代码实现
- [置顶] 【深度学习】RNN循环神经网络Python简单实现
- [深度学习]Python/Theano实现逻辑回归网络的代码分析
- 神经网络与深度学习 1.6 使用Python实现基于梯度下降算法的神经网络和MNIST数据集的手写数字分类程序
- 深度学习第一课 第四周 深层神经网络用python的实现
